| AlphaFold DeepMind's Protein Structure Breakthrough |
| Written by Mike James | |||
| Wednesday, 12 December 2018 | |||
|
AlphaGo learned to play Go at grand-master level. AlphaFold has used similar methods to learn to predict protein folding and this is far more important. Protein folding sounds like a strange sort of problem. What it is all about is that DNA sets out the sequence of amino acids that have to be joined together to build a protein, but it only determines their order. Once the protein chain has been formed it snaps into a 3D shape - a bit like a straight piece of wire that has previously been bent into a shape that it "remembers". The way a protein folds to create the 3D shape is determined in a complex way by the attractions between the different amino acids. The protein twists in space so as to bring strongly attracting amino acids close together while minimizing the local bending of the structure. If you have a sequence of amino acids, as say read off from a DNA sequence, then this gives you the chemical formula of the protein but not its all important 3D shape. Working out its 3D shape is the protein folding problem and it is very difficult. You can probably guess that solving the protein folding problem has lots of real applications and we really do want to work out how to do it. Conventional optimization procedures are far too slow and this is where neural networks come into the picture. DeepMind joined in a competition to predict protein folding with AlphaFold, a neural network. It was placed first and was a lot better than its closest rival.
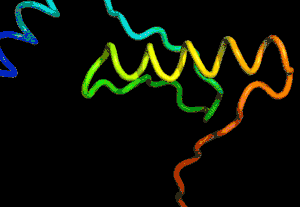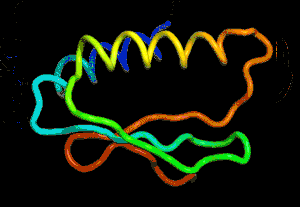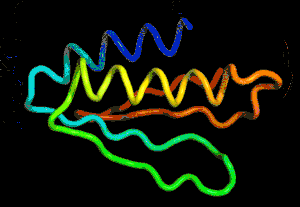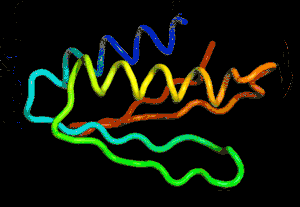
After the physical properties have been worked out data fragments of known protein structure were fitted to give a 3D shape that produced the same properties. A generative neural network was also trained to generate new protein fragments which were also used to improve the shape. A second method attempted to fold the entire chain to fit the predictions of distance and angle using optimization - see the animation below. A simple gradient descent was used to bend the chain so as to meet the predicted physical properties. If you look carefully you can see the the intial folding is quite fast and then the gradient descent slowly "tinkers" with the fine adjustment to improve the fit.
Animation of the gradient descent method of predicting the structure of a folded protein. This is not an obvious application of AI, but it is the sort of thing that could be revolutionary in many fields in the future. AI gives us the ability to solve problems in new ways - the only problem is we don't know how much to trust the results as AI predictions rarely come with error bounds. It also seems that the other competitors and onlookers were a bit shocked that an AI team could "gate crash" their party and take over a field of study without really knowing anything about it. This is likely to happen increasingly often in AI lead science. More InformationAlphaFold: Using AI for scientific discovery Related ArticlesNobel Prize For Computer Chemists A Neural Network Reads Your Mind AlphaGo The Movie - Now On Netflix To be informed about new articles on I Programmer, sign up for our weekly newsletter, subscribe to the RSS feed and follow us on Twitter, Facebook or Linkedin.
Comments
or email your comment to: comments@i-programmer.info |
|||
| Last Updated ( Wednesday, 01 April 2020 ) |
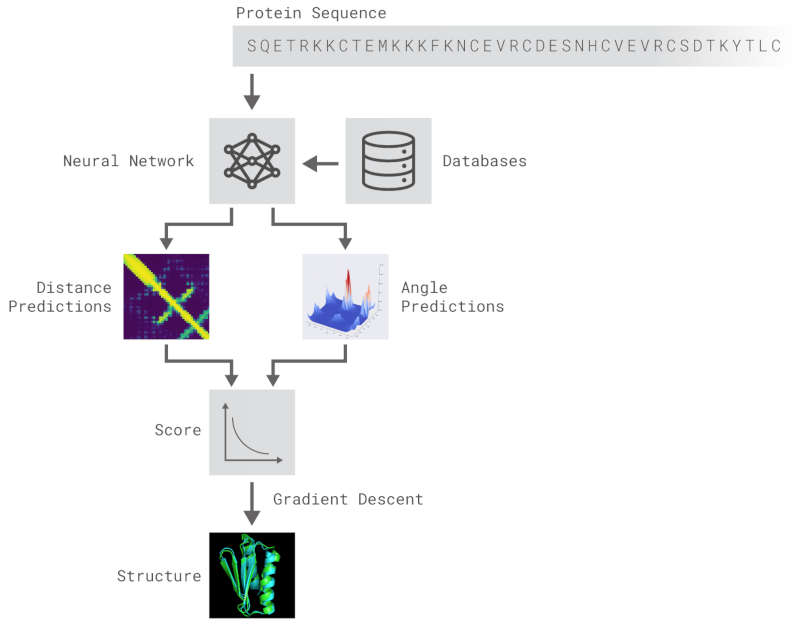 It performed the feat in a number of steps. The first seems an unlikely achievement - the neural network learned to predict properties such as the distances between pairs of amino acids and the angles between bonds from just the DNA sequence.It seems unlikely that you can predict such things from just a linear amino acid sequence but it seems you can.
It performed the feat in a number of steps. The first seems an unlikely achievement - the neural network learned to predict properties such as the distances between pairs of amino acids and the angles between bonds from just the DNA sequence.It seems unlikely that you can predict such things from just a linear amino acid sequence but it seems you can.