| Genomics Needs Better Compression |
| Written by Mike James | |||
| Wednesday, 29 August 2018 | |||
|
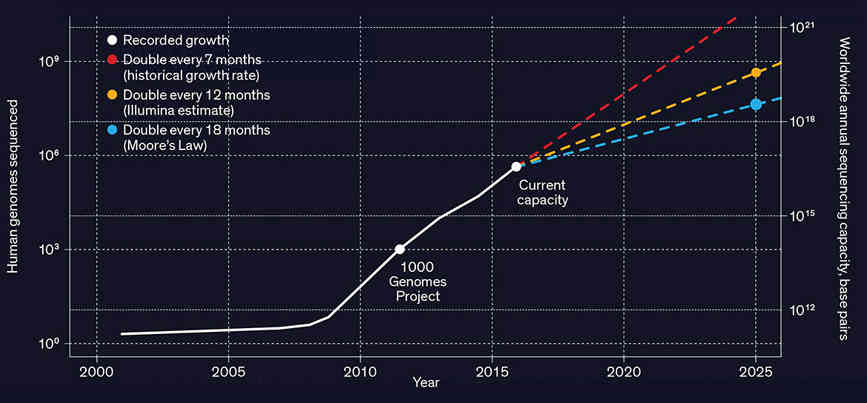
An article in the IEEE Spectrum highlights a problem that, by my guess, few of us are aware of. The simple fact is that genetics is generating data at such a rate that, without improved compression algorithms, the subject will not deliver on its promise. Before we get started on the serious stuff, it is interesting to comment that the authors of the article, Dmitri Pavlichin and Tsachy Weissman, are familiar to a wider audience because they were both consultants to the TV show Silicon Valley. They advised on the invention of a compression algorithm that the startup was supposed to be exploring. In fact both of them are trying to do something similar in real life, but with the focus on genomics. First we need to understand the size of the problem. Genomic sequencing of a human generates tens to hundreds of gigabytes of data. This is manageable but, as gene sequencing gets cheaper, it is becoming thinkable to sequence the entire population and not just once but multiple times to see how things change. We are talking exabytes of data and the need to process it.
Source: Stephens ZD, Lee SY, Faghri F, Campbell RH, Zhai C, Efron MJ, et al., 2015, PLoS Biol 13(7) What is needed is a compression algorithm tailored to genomic data. The problem is made worse by the fact that the data is very fragmented. A sequencing machine doesn't produce a long string of ACGT coding a single genome but a large set of overlapping fragments that have to be assembled. The data also contains errors and sequencing machines output a quality score to indicate how sure they are of the data. All of these fragments and quality scores are output in a single FASTQ file which is up to 100 Gigabytes. The good news is that while there is a lot of data there is a lot of redundancy due to the resequencing of specific portions of the DNA to check for errors. You could use a general compression algorithm, such as ZIP or one of the recent modifications of dictionary compression, but it seems more likely that a genome-specific method would fit in with the sort of processing required.
For example, one approach is to use a single genome sequence as a reference dictionary and then describe the sequences you have in terms of starting position and any deviations from the standard. This sounds good, but it doesn't deal with the quality scores. There is also work to be done in characterizing the statistics of DNA. If you recall, all compression relies on using probabilistic patterns that reduce the entropy of the data from uncompressible random data to something more regular and compressible. The authors of the article report a double power law that they have found which puts some regularity on the dispersion of genetic variations. This is an interesting finding in itself from the point of view of what aspect of evolution causes it, but it can also be used to fine tune compression algorithms. At the moment all genomic compression algorithms are lossless and it is an open question if lossy algorithms might be good enough - not so much for the DNA sequence, but for the quality scores which take a lot of bits to store. This is an area where we need new ideas and new ways of processing the growing mountain of data. Clearly, standards would help future work and the strange fact is that the Moving Picture Experts Group - MPEG - is the body working on creating a standard. MPEG-G should be finished sometime before the end fo the year. The last word should go to the authors: "Genomic researchers have started applying deep learning to their data, but they’ll likely have to wait for a critical mass of genomic information to accumulate before realizing similar gains. One thing is clear, though: They won’t get there without major advances in genomic data compression technology."
More InformationThe Desperate Quest for Genomic Compression Algorithms Related ArticlesTraveling Salesman Applied To DNA Synthesis Book Stored On DNA - All Knowledge In Just 4gm of DNA A New DNA Sequence Search - Compressive Genomics Coding Contest Outperforms Megablast E. coli Your Next Major Platform Programmed In Cello Edit Distance Algorithm Is Optimal Genetic Algorithms Reverse Resistance To Antibiotics To be informed about new articles on I Programmer, sign up for our weekly newsletter, subscribe to the RSS feed and follow us on Twitter, Facebook or Linkedin.
Comments
or email your comment to: comments@i-programmer.info The Desperate Quest for Genomic Compression Algorithms |
|||
| Last Updated ( Wednesday, 29 August 2018 ) |