| AlphaFold Reads The DNA |
| Written by Mike James | |||
| Wednesday, 28 July 2021 | |||
|
We have had the map of the human genome for a while, but only now can we read it. Alphafold, DeepMind's most useful AI to date can work out what the DNA is actually saying and DeepFold source code has been released.
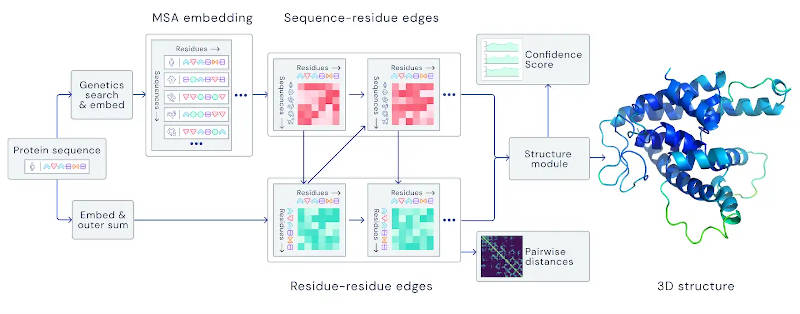
AlphaFold solves a problem that doesn't really seem to be mainstream AI - it is more like chemistry or physics - but the impact of AI on science is likely to increase in the near future. The problem is simple to state and this makes it all the harder to appreciate how difficult it is to solve. We have the sequence of bases that make up human DNA and this forms a set of instructions how to make proteins, which is what biological systems are made of. The problem is that the DNA gives only the order that the different amino acids are assembled in. When they are made they form a linear string of amino acids that only form a 3D shape once released from the DNA factory. The linear string very quickly folds itself up into a shape that essentially determines its physical properties. So to be able to predict what each portion of the human genome actually does, we need not only to know how the base pairs translate into amino acid sequences, which is easy, but how the protein will fold when the construction work is finished and this last part if very difficult. The traditional way of doing the job is essentially to synthesize the protein in the lab, let it fold and then perform x-ray crystallography to get its 3D structure. This is very slow. A faster method would be to let AI solve the problem and this is what DeepMind, and many others, have been working on. AlphaFold was announced last year and now we have details of how it works. However, over the same time period a team at the University of Washington, RoseTTAFold, which claims to be inspired by AlphaFold, has come up with a method that is more efficient and just as accurate. This development might well have made DeepMind speed up the open sourcing of AlphaFold 2. DeepMind used the equivalent of 200 GPUs to train and uses a parallel approach to the problem that overall isn't that revolutionary. That is, the breakthrough is in chemistry rather than AI. One process looks at already-known structures that are similar to the one being constructed and then comes up with a proposed structure. A second process looks are smaller chunks of the amino acid chain to find sub-units that are compatible with the whole structure. The two processes make use of each others results to refine the overall structure.
So we now have a tool to decode the DNA what next? One week after releasing the information on how AlphaFold works, DeepMind announced that it was planning to let the program work on the entire human genome so creating an open source database of proteins. Beyond this it has plans to extend the work to an additional 20 important organisms making the projected size of the database roughly 300,000 proteins. Given that at the moment we only know the structure of less than 20% of human proteins, this represents a major step forward. Even so there are still "big" proteins that have too many amino acids to compute and anything larger than 2700 amino acids will be left out, giving an overall 98% coverage of the genome. It is clear that AI and exceptionally big data are both going to play a role in our understanding of biology and our application of that understanding to medicine. Now you know what terabyte disks and GPUs where invented for. More InformationHighly accurate protein structure prediction with AlphaFold https://github.com/deepmind/alphafold AlphaFold Protein Structure Database Accurate protein structure prediction accessible to all Related ArticlesAlphaFold Solves Fundamental Biology Problem AlphaFold DeepMind's Protein Structure Breakthrough Fighting Coronavirus At Home With Exascale Power Nobel Prize For Computer Chemists To be informed about new articles on I Programmer, sign up for our weekly newsletter, subscribe to the RSS feed and follow us on Twitter, Facebook or Linkedin.
Comments
or email your comment to: comments@i-programmer.info |
|||
| Last Updated ( Wednesday, 28 July 2021 ) |