| Traveling Salesman Applied To DNA Synthesis |
| Written by MIke James |
| Wednesday, 13 January 2016 |
|
One of the fun things about being a programmer is that algorithms that you know can often be applied in unexpected ways in areas that are the domain of other specialists. A good example is the traveling salesman problem being applied to DNA synthesis. If you want to create a custom strand of DNA, it is fairly automatic these days. You specify the sequence of base pairs you need and a machine can create the DNA for you. Automatic DNA or gene synthesis works by using the standard Polymerase Chain Reaction (PCR) that is used in genetic "fingerprinting". In this process smaller sections of DNA are joined together and reproduced until a usable quantity has been created. This works well, but if the desired DNA contains repeated sections then it fails because there are multiple ways that the DNA fragments can be assembled. This seems to be a fundamental problem with the method, but if you are only interested in the protein that the DNA fragment codes for then you have some extra degrees of freedom. The protein that the DNA codes for is a sequence of amino acids and every three letters of the DNA, a codon, specifies a single amino acid, but there are 61 codons producing only 20 amino acids. What this means is that you can change the DNA sequence by substituting equivalent codons and still use it to generate the same protein. The redundancy in the codons is the key to solving the repeated sequence problem. All we have to do is change the codons to remove the repeats in the DNA while preserving the repeats in the protein produced. This is what scientists from Duke University have done using an analogy with the traveling salesman problem. The task is to find the least repetitive sequence that still produces the same protein. Ashutosh Chilkoti, the Theo Pilkington Professor of Biomedical Engineering explains: "I always thought there was a potential solution, that there must be a way of mathematically figuring it out. I had offered this problem to graduate students before, but nobody wanted to tackle it because it requires a particular combination of high-level math, computer science and molecular biology. But Nicholas Tang was the right guy." The solution is a version of the "traveling salesman" problem. The classic question is, given a map with a set of cities to visit, what is the shortest route possible that hits every city exactly once before returning to the original city? Once recognized as the traveling salesman problem an exact solution using De Bruijn graphs and a modern mixed integer linear programme solver was found.
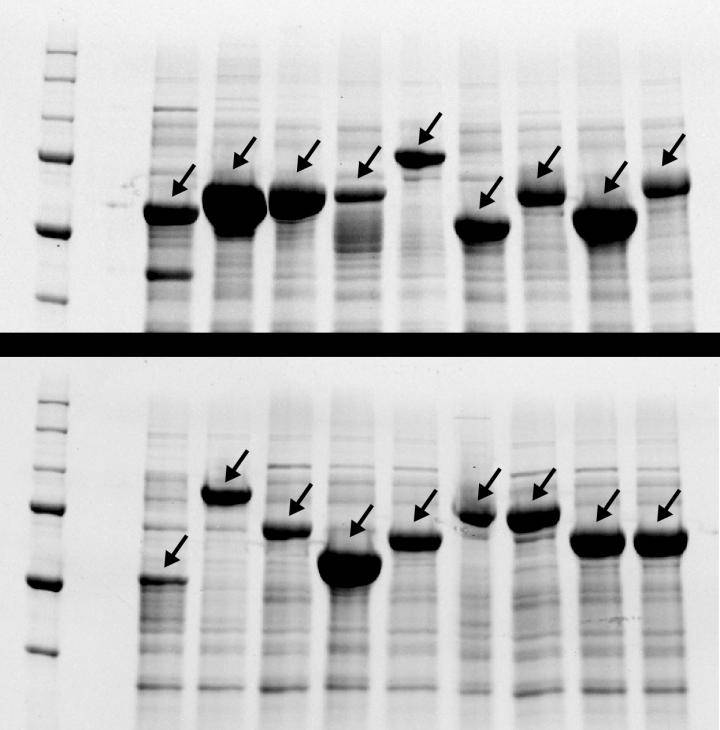
After swapping pieces of genetic code in repeating protein recipes, researchers were able to scramble them enough to be commercially synthesized. The dark bands are indicators of 18 repeating polypeptides successfully being fabricated. To prove that he had a solution, Tang made non-repetitive DNA for 19 highly repetitive polypeptides and sent the specifications off to a commercial lab. They returned working samples of the DNA with no problems. This would have been impossible using the original DNA specifications. Chilkoti and Tang are now working to make the new computer program available online for anybody to use through a simple web form, opening a new area of synthetic biology for all to explore.
More InformationCombinatorial codon scrambling enables scalable gene synthesis and amplification of repetitive proteins Nicholas C. Tang & Ashutosh Chilkoti (paywalled) Related ArticlesCoding Contest Outperforms Megablast Book Stored On DNA - All Knowledge In Just 4gm of DNA A New DNA Sequence Search - Compressive Genomics
To be informed about new articles on I Programmer, sign up for our weekly newsletter, subscribe to the RSS feed and follow us on, Twitter, Facebook, Google+ or Linkedin.
Comments
or email your comment to: comments@i-programmer.info
|
| Last Updated ( Wednesday, 13 January 2016 ) |