| Coding Contest Outperforms Megablast |
| Written by Sue Gee |
| Wednesday, 13 February 2013 |
|
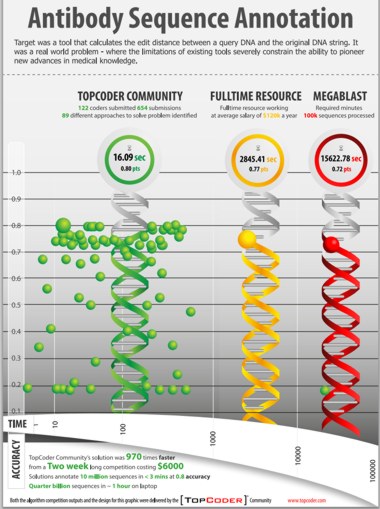
A $6,000 coding contest to solve a "big data" problem in computational biology produced a solution that was 970 times faster than existing solutions. This spectacular success story tells how TopCoder, a community with more than 450,000 members representing "algorithmists", software developers and creative artists from over 200 countries, came up with a range of innovative approaches to the real world problem of analyzing the sequence data from the genes and gene mutations that build antibodies and T cell receptors. The challenge for a 2-week contest with three weekly prizes of $500, was to develop a predictive algorithm that would outperform both the NIH's standard approach (MegaBLAST) and an alternative custom algorithm developed by Ramy Arnaout of the Beth Israel Deaconess Medical Centre. In setting the challenge, the researchers, led by Eva Guinan , HMS associate professor of radiation oncology at Dana-Farber Cancer Institute, reframed the problem to make it accessible to individuals not trained in computational biology in terms of finding the edit distance between a query DNA and the original DNA string.
(click on poster to view it in new window) Source: TopCoder.com
The bottom line, as summarized in the poster, was that the TopCoder community came up with 654 submissions and provided 89 different approaches to solving the problem. 16 solutions were an improvement over MegaBLAST with one over 970 times faster than either it or the HMS algorithm. More details are provided in TopCoder's press release: The challenge drew 733 participants, of whom 122 (17%) submitted software code. This group of submitters, drawn from 69 countries, included roughly half (44%) professionals with the remainder being students at various levels. None were academic or industrial computational biologists, and only five described themselves as coming from either R&D or life sciences in any capacity. The 122 TopCoder members submitted 654 submissions yielding 89 different approaches to the problem. Collectively, participants averaged 5.4 submissions each. Participants reported spending an average of 22 hours developing solutions, for a total of 2,684 hours of development time. Sixteen of the submissions outperformed the accuracy (77%) of the traditionally developed custom solution and 30 outperformed the NIH MegaBLAST benchmark for accuracy (72%). A total of eight submissions achieved an 80% accuracy score, which is very near the theoretical maximum for the dataset. The February 7, 2013 issue of Nature Biotechnology includes an article from the team of the researchers and its title proclaims the conclusion: "Prize-based contests can provide solutions to computational biology problems".
More InformationRelated ArticlesA New DNA Sequence Search - Compressive Genomics Book Stored On DNA - All Knowledge In Just 4gm of DNA
To be informed about new articles on I Programmer, install the I Programmer Toolbar, subscribe to the RSS feed, follow us on, Twitter, Facebook, Google+ or Linkedin, or sign up for our weekly newsletter.
Comments
or email your comment to: comments@i-programmer.info
|
| Last Updated ( Wednesday, 13 February 2013 ) |